Test phylogenetic signal
19 Dec 2015Phylogenetic signal
Ecological traits of related species always tend to be more similar than species randomly drawn from the same phylogeny. This is called phlogenetic signal[1-3]. It is often interpreted as providing information about the evolutionary process, and is controlled as the statistical nonindependence among species trait values in ecological analysis. There are some posts about how to test phylogenetic signal in R, this, this and this. I would give another example which I was recently involved in.
Steps in R
Let’s take some amphibians with a virtual trait for example.
0. Amphibians with a virtual trait
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
species <- read.csv("amphibians.csv", stringsAsFactors = F)
# Some amphibian species.
# Variable "tip_labels" is the species' name in the phylogenetic tree,
# while "species_name" is the name I got.
# They are the synonyms, or they are same.
# I retain them two for demonstrating
# how to substitute the tip labels later.
species
## species_name tip_labels
## 1 Alytes_obstetricans Alytes_obstetricans
## 2 Ambystoma_tigrinum Ambystoma_tigrinum
## 3 Amietophrynus_regularis Bufo_regularis
## 4 Anaxyrus_americanus Bufo_americanus
## 5 Aneides_vagrans Aneides_vagrans
## 6 Dendrobates_auratus Dendrobates_auratus
## 7 Desmognathus_quadramaculatus Desmognathus_quadramaculatus
## 8 Discoglossus_pictus Discoglossus_pictus
## 9 Duttaphrynus_melanostictus Bufo_melanostictus
## 10 Eleutherodactylus_coqui Eleutherodactylus_coqui
## 11 Eleutherodactylus_johnstonei Eleutherodactylus_johnstonei
## 12 Eleutherodactylus_planirostris Eleutherodactylus_planirostris
## 13 Glandirana_rugosa Rana_rugosa
## 14 Hoplobatrachus_rugulosus Hoplobatrachus_rugulosus
## 15 Hoplobatrachus_tigerinus Hoplobatrachus_tigerinus
# A virtual variable
set.seed(1234)
species$x <- rnorm(dim(species)[1], mean = 50, sd = 10)
1. Built the phylogenetic tree
Actually prune a built tree from Pyron & Wiens (2011)[4].
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
# Load library
library(ape)
supertree <- read.tree("amphibia.tre")
# Remove tips and branches that will not be used
tree_am <- drop.tip(supertree,supertree$tip.label[!(supertree$tip.label
%in% species[,"tip_labels"])])
# Substitute the tip labels (tip_labels) with the labels I want (species_name)
matched_labels <- tapply(tree_am$tip.label,
1:length(tree_am$tip.label),
function(x) species[which(species$tip_labels == x), "species_name"])
tree_am$tip.label <- as.data.frame(matched_labels)[,1]
# Plot the tree (Fig. 1)
plot(tree_am)
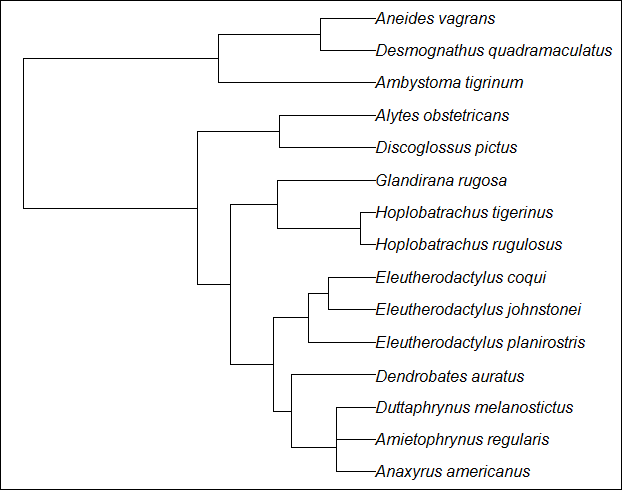 Figure 1 Amphibian phylogeny.
Figure 1 Amphibian phylogeny.
2. Test phylogenetic signal
Use the indices of Pagel’s lambda and Blomberg’s K. See Pagel (1999)[5] and Blomberg, et al. (2003)[6] for details.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
# Load library
library(phytools)
# Named the variable
x <- species$x
names(x) <- species[,"species_name"]
phylosig(tree_am,x,method="lambda",test=T)
## $lambda
## [1] 6.901966e-05
## $logL
## [1] -53.78967
## $logL0
## [1] -53.78936
## $P
## [1] 1
phylosig(tree_am,x,method="K",test=T)
## $K
## [1] 0.2171225
## $P
## [1] 0.811
No phylogenetic signal was detected.
References:
[2]: Hof, C., Rahbek, C. & Araujo, M.B. (2010) Phylogenetic signals in the climatic niches of the world's amphibians. Ecography, 33, 242-250.
[3]: Münkemüller, T., Lavergne, S., Bzeznik, B., Dray, S., Jombart, T., Schiffers, K. & Thuiller, W. (2012) How to measure and test phylogenetic signal. Methods in Ecology and Evolution, 3, 743-756.
[4]: Pyron, R.A. & Wiens, J.J. (2011) A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Molecular Phylogenetics and Evolution, 61, 543-583.
[5]: Pagel, M. (1999) Inferring the historical patterns of biological evolution. Nature, 401, 877-884.
[6]: Blomberg, S.P., Garland, T., Jr. & Ives, A.R. (2003) Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution, 57, 717-745.